![]()
Center for Neuroscience and
Department of Anatomy and Neurobiology
University of Tennessee
855 Monroe Avenue
Memphis Tennessee 38163, USA
email rwilliam@nb.utmem.edu
Common genetic polymorphisms—as opposed to rare mutations—generate almost all heritable differences in the size and structure of the CNS. Surprisingly, these normal variants have not previously been mapped or cloned in any vertebrate species. In a recent paper (Williams et al., 1996a) we suggested that much of the variation in retinal ganglion cell number in mice, and the striking bimodality of strain averages, is caused by one or two quantitative trait loci (QTLs). To test this idea, and to map genes linked to this variable and highly heritable quantitative trait, we have counted ganglion cells in 38 recombinant inbred strains (BXD and BXH) derived from parental strains that have high and low cell numbers. A genome-wide search using simple and composite interval mapping techniques revealed a major QTL on chromosome (Chr) 11 in a 3 cM interval between Hoxb and Krt1 (LOD = 6.8, genome-wide P < 0.001), and possible subsidiary QTLs on chromosomes 2 and 8. The Chr 11 locus, neuron number control 1 (Nnc1), accounts for one-third of the genetic variance among BXH strains and more than half of that among BXD strains, but Nnc1 has no known effects on brain weight, eye weight, or total retinal cell number. Three strong candidate genes have been mapped previously to the same region as Nnc1. These genes—Rara, Thra, and Erbb2—encode receptors for retinoic acid, thyroxine, and neuregulin, respectively. Each receptor is expressed in the retina during development, and their ligands affect the proliferation or survival of retinal cells.
The most conspicuous differences between the brains of different mammalian species are quantitative (2, 3). Total brain weight, the size of different brain nuclei, and numbers of neurons can vary over two or three orders of magnitude (4). This marked variation ultimately traces back to differences that are generated, selected, and propagated within single species. Two impressive examples of variation in the human CNS include the three-fold difference in the density of cone photoreceptors in the fovea (5) and the three-fold differences in the total area of both primary and secondary visual cortex (6). Some of this variation is undoubtedly environmental, but much is generated by the independent segregation of alleles that control proliferation, migration, differentiation, and survival of neurons and glial cells. None of the genes that are responsible for this normal variation in the mammalian CNS structure have yet been mapped or identified. Yet these genes are particularly important because they ultimately influence the performance and behavioral repertoire of a species.
Genetic variation in complex traits is thought to be generated by large numbers of loci that generally have comparatively small effects (7). However, a subset of these loci have surprisingly large individual effects. For example, single quantitative trait loci (QTLs) have been shown to account for 20-40%; of the variance in the height of corn and the weight of tomatoes (8). Similarly, several QTLs individually account for as much as 10%-20%; of the total variance in numbers of sensory bristles in fruit flies. Some of these QTLs are now known to correspond to key neurogenic or proneural genes, including achaete-scute, atonal, enhancer of split, hairy, Notch, and scabrous (9).
To map genes that contribute to normal variation in the vertebrate CNS, we have focused on an important and well-defined class of sensory neurons called retinal ganglion cells. Axons of these neurons give rise to the optic nerve and are essential for transmitting visual information to the thalamus and midbrain. Variation in ganglion cell number is generated primarily by genetic factors (heritability is approximately 0.8) (1). The distribution of ganglion cell number is close to normal, with a mean of 60,000 and a range from 40,000 to 80,000. In this respect, variation in ganglion cell number is a typical complex trait displaying continuous variation over a wide range. However, one surprising finding from our previous work is that the distribution of inbred strain averages—as opposed to individual values—is distinctly bimodal, with modes near 55,500 and 63,500 (1). This pattern could be generated by the segregation of high and low alleles at a major QTL. In this study we have mapped this QTL to a gene-rich region on Chr 11 between Hoxb and Krt1. This is the first locus known to control normal variation in CNS structure in a vertebrate.
Fixation and processing of tissue. Eyes, optic nerves, and brains were taken from 182 cases belonging to the 26 BXD strains, 66 cases belonging to the 12 BXH strains, and 45 cases from the three parental strains. All mice were obtained from the Jackson Laboratory, Bar Harbor, Maine. Mice of both sexes and a wide range of age (30 to 400 days) were anesthetized with an injection of avertin (0.5–0.8 ml ip) and were perfused transcardially with phosphate buffered saline followed by 1.25%; glutaraldehyde and 1.0%; paraformaldehyde in 0.1 M phosphate buffer, and then by 2.5%; glutaraldehyde and 2.0%; paraformaldehyde in 0.1 M PB. Nerves were dissected, osmicated, and embedded in Spurr's resin. Brains, including olfactory bulb, were dissected, and weighed. Thin sections of one or both nerves were placed on formvar-coated slot grids and were stained with uranyl acetate and lead citrate. The nerves were examined and photographed on an electron microscope using a systematic sampling protocol1.
C3H/HeJ and 8 of the BXH strains (2, 3, 4, 7, 8, 9, 14, and 16) are all homozygous for the photoreceptor degeneration allele, rd1, at the beta phosphodiesterase locus on Chr 5. Despite a massive loss of photoreceptors during the first two months of life, this mutation has no detectible effect on the retinal ganglion cell population (r = –0.3). The eight strains that have the rd1 allele have 61,500 +/- 2000 (SE) ganglion cells, whereas the four strains that have the wildtype allele have 58,300 +/- 4400 (SE) ganglion cells.
Phenotyping. Each ganglion cell has a single axon that extends from the retina into the brain1, 53. It is therefore possible to count this cell population simply by counting nerve fibers in single cross-sections of the optic nerve. A counting frame was traced on negatives with a marker and all axons within the frame and intersecting the upper and right edges were marked and counted on the negative using stereological counting rules. To ensure that unmyelinated fibers were recognized, negatives were counted while wearing magnifying glasses. The effective magnification was above X25,000. Approximately 90 cases were independently replicated. All data were entered into a spreadsheet (Excel 5.0, Microsoft), available at http://www.nervenet.org/main/maincontrol.html. The average density of axons was multiplied by the area of the nerve cross-section to estimate the total axon population. Strain averages are presented as unweighted means. To perform linear regression and compute residuals we used the program DataDesk 5.0 (Data Description). All individual brain weights were initially adjusted to that expected of females weighing 22 gm (54). This improves the quality of comparisons across strains. A regression analysis of the stain averages of brain weight and neuron number was then carried out to compute the residuals listed in Tables 1 and 2.
Analysis of retinas. A 1-mm-wide strip of retina and pigment epithelium, extending from the head of the optic nerve to the inferior ora was cut out of one eye from each of 16 cases and embedded flat in Spurr's resin. The 1-µm-thick sections were cut along the radial axis, mounted, and stained with hematoxylin. Slides were coded, but were otherwise left unlabeled. Complete central-to-peripheral cross-sections of the ventral retina were drawn at low power. The radial depth of cells in the inner and outer nuclear layers was subsequently determined at 7 to 11 evenly spaced sites along all sections at X400 using differential interference contrast optics. Ambiguity of these counts at single sites is less that ± 2 cells. The outer nuclear layer is between 5 and 15 cells deep, whereas the inner layer is between 2 and 6 cells deep. The average coefficient of variation within a case for these measurements was 5.8%; in the outer layer and 7.5%; in the inner layer.
QTL mapping. Simple and composite interval mapping (55-57) was performed using the program Map Manager QT58 and a dataset of RI strain genotypes compiled by R. W. Elliott and B. Taylor. The original BXD file is comprehensive and contains many groups of linked loci that have identical strain distribution patterns. All uninformative and incompletely typed loci, and loci with numerous unexplained double recombinants over short intervals were deleted from the dataset. The final BXD database used for interval mapping and permutation analysis contained 580 completely typed loci that defines a genome with a total length of approximately 1,640 cM. Genome-wide signficance was estimated by comparing the peak likelihood ratio statistic (LRS = 4.6 x Lod) of the correctly ordered data with those computed for 1,000 to 10,000 random permutations of the data59. To combine probabilities from the two RI sets we computed the probability associated with a chi-square value equal to -2* (lnPBXD+lnPBXH) with 4 degrees of freedom, where lnPBXD and lnPBXH are the natural logarithms of the probabilities derived independently for the two RI sets. We have mapped both the average number of retinal ganglion cells and the residual number of ganglion cells after eliminating effects of variation in sex, body weight, brain weight, and age. Files used for mapping are available at http://www.nervenet.org/main/maincontrol.html. Mapping data have been deposited with the Mouse Genome Database.
Two groups of recombinant inbred strains were studied—the set of 26 BXD strains generated by crossing C57BL/6J to DBA/2J mice, and the set of 12 BXH strains generated by crossing C57BL/6J to C3H/HeJ mice. The maternal strain, C57BL/6J, is common to both BXD and BXH RI sets. C57BL/6J (or B for its black coat color) belongs to the low-cell-number group and has a population of 55,400 ± 800 ganglion cells (n = 26). Both paternal strains, DBA/2J (D) in the case of the BXD RI set and C3H/HeJ (H) in the case of the BXH set, belong to the high group and have populations of 63,400 ± 1,200 (n = 13) and 67,000 ± 1,700 (n = 6), respectively.
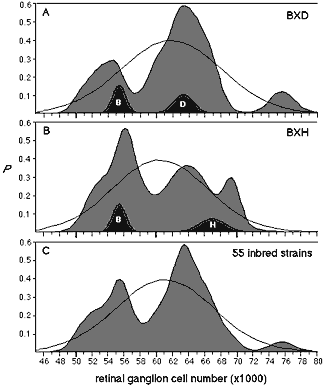
Figure 1. Probability density of strain averages for the BXD RI strains (bold curve) suggests that there are three relatively distinct phenotypes. A goodness-of-fit test confirms that the distribution not normal (P < 0.05). The small functions labeled B and D are Gaussian probability density functions of the parental strains. A set of 26 similar functions for BXD strains were added to generate the summed probability density for the entire BXD series. Original data are listed in Table 1. The expected Gaussian function is drawn in lightly. b, Probability density function for the 12 BXH strains (bold curve) and the two parental strains. c, A summed probability density function for all 55 inbred strains of mice, including the 38 RI strains listed in this paper and 17 standard inbred strains listed in Ref. 1. This large collection is less subject to sampling error and provides our best estimate of the effect of allelic substitutions at Nnc1. F1 hybrids between high and low strains typically have intermediate cell populations between 56,000 and 64,000 (average 61,500, n = 7; Ref 1).
The average ganglion cell population in the BXD strains extends from a low of 50,800 ± 1,100 in BXD27 to a high of 75,800 ± 2,000 in BXD32 (Table 1). The probability density function for the 26 BXD strains has modes at 54,000 and 64,000 (1a). These modes correspond almost precisely to the means of the parental strains and are aligned with the prominent modes discovered in our previous analysis of 17 common inbred strains (see Fig. 4 of Ref. 1). Four strains—BXD1, BXD11, BXD20, and BXD21—have averages that are in the central range (59,900 to 61,000), reasonably close to the midparental value of 59,000. Strains BXD5 and BXD32, both of which have very high cell number, represent a third mode. Average cell number in the set of BXH RI strains extends from 51,000 to 70,000, without significant transgression above or below the parental values (Table 2). The probability density function for these strains is broad and is characterized by a prominent peak at approximately 56,000 and poorly resolved modes at 64,000 and 70,000 (Fig. 1b).
In fully homozygous RI lines the independent assortment of n diallelic loci gives rise to a maximum of 2n genotypes (e.g., if n = 2 then the genotypes are aabb, aaBB, AAbb, AABB), and each genotype is expected to be represented equally in the set of RI strains. If a large number of loci with intermediate or small additive effects assort independently during the generation of BXD and BXH strains, the probability densities in figure 1 would tend to have a unimodal and perhaps Gaussian form. In contrast, the broad and multimodal distributions of the RI sets, and the clearly bimodal distribution of the data based on all 55 inbred strains (Fig. 1c), suggest that a small number of QTLs have major effects on neuron number (note 1). This is consistent with our finds in F2 intercrosses, in which the number of effective factors controlling ganglion cell number has been found to be less than three (10). A comparision of variance within and among these isogenic strains indicates that genetic factors account for approximately 70%; ± 10%; of the total phenotypic variance in the RI strains (1, 11).
The distribution of phenotypes listed in Tables 1 and 2 can be compared directly to those loci that have already been mapped by hundreds of investigators over the past decade. The single best match on the densely mapped set of BXD strains is to the tissue specific transplantation antigen 91A gene (Tstap91A) located approximately 2 cM distal to Hoxb on Chr 11 (12). Seven of the strains that have low cell number correspond to the C57BL/6J alleles (B for short) at Tstap91A, and 15 that have high cell number correspond to DBA/2J alleles (D for short). The four remaining strains with intermediate phenotypes correspond to 2 B and 2 D genotypes at Tstap91A. Only one strain, BXD31, is unequivocally discordant. The correlation coefficient between neuron number and alleles at Tstap91A is 0.69. (To compute this correlation, B alleles at Mendelian loci are arbitrarily assigned a value of 0, and D alleles are assigned a value of 1.) The Lod score for linkage of retinal ganglion cell number with Tstap91A is 3.7 (Table 3). The probability of achieving this statistic by chance is 0.000037 for a test against a single marker and 0.06 for multiple tests covering the entire genome.
These values underestimate the actual strength of linkage because variation in brain weight obscures variation in neuron number associated with the Chr 11 QTL. This global influence can be minimized by computing residuals after regressing cell number against brain weight (Table 1, right-most column). Mapping these residuals results in a significant improvement in the strength of linkage (Lod 4.4, single-point P = 0.0000065, genome-wide P <0.05). Two other chromosomal intervals—one on Chr 2, the other on Chr 8—were shown to be well correlated with the remaining genetic variation in ganglion cell number (see below). We subsequently corrected for the effects of these two intervals and for that of a third interval on proximal Chr 11 near Glns-ps1 that has been previously shown to have a significant effect on brain weight among BXD strains13. In the final analysis the Lod for linkage between cell number and the Tstap91A interval is approximately 6.8 (Fig. 2, P = 2.0 x 10 E -8, genome-wide P < 0.001). We have named this major effect QTL on Chr 11 Neuron number control 1 (Nnc1). Nnc1 maps between the Hoxb complex and Mpmv8, an interval of approximately 3 cM (Fig. 2). The probability of linkage drops more than 100-fold outside of this interval. Independent support for linkage is provided by the BXH strain data, in which one of the strongest associations between H alleles and strains with high cell population (r = +0.58) is also on mid-distal Chr 11 between Scya and Krt1 (P = 0.01).
An analysis of the 12 BXH strains shows that much of the variation in neuron number in this RI set could be accounted for by a QTL on Chr 4. The correlation between alleles at Ssdh1 on Chr 4 and cell number is tight but negative (r = -0.92; Lod is 4.8; P = 2.8 x 10 E-6). Similar statistics are obtained when mapping residuals that control for the negative correlation between brain weight and neuron number. Despite these persuasive statistics, we have good reasons to believe that this linkage is spurious. First, B alleles inherited from the parental strain with low cell number are consistently associated with RI strains with high cell number. Such a reversal, while not uncommon in mapping QTLs that have modest effects, is unexpected for a QTL with such a massive apparent effect (r2 = 0.8). Perhaps more telling is the observation that the strain distribution pattern at Ssdh1 on Chr 4 is almost precisely the same as that of Rasl5-2 on Chr 4 (11 of 12 common alleles), and nearly opposite to that of Scya3 (10 of 12 reversed alleles)—a locus within 10 cM of Tstap91A on Chr 11. Given the small number of BXH strains and the high density of typed loci, false associations are common. The set of 12 BXH strains is too small to map loci with any confidence, but data from small RI sets can be combined to add power and resolution to a QTL analysis.
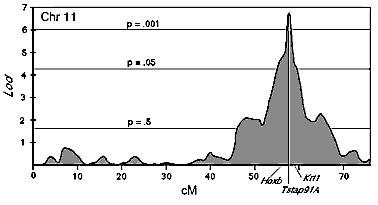
After controlling for variance associated with Nnc1 on Chr 11, two additional intervals that may contain QTLs that modulate cell number were highlighted in both RI sets. The first is located near Lpl and Cpe on Chr 8 (approximately 33 cM). The combined Lod for both RI sets in this region is 2.8 (single-point P = 0.0004). The second interval is between B2m and Mltr10 on Chr 2 and has a combined Lod of 2.4. (single-point P = 0.001 at Hdc). Clearly, the statistics are not strong enough to claim QTLs in either interval. As mentioned above, both regions were subsequently taken into account in composite interval mapping of the Chr 11 locus. This resulted in a significant improvement in linkage to the Tstap91A interval on Chr 11. Collectively as much as 70%; of the variation in neuron number and as much as half of the total phenotypic variance can be accounted for by Nnc1 and by putative secondary QTLs on Chrs 2 and 8.
The phenotypic effects of alleles at single loci are difficult to estimate from sets of RI strains, and estimates tend to be much too high (14). However, in this case, the clear separation between high and low strains, shown particularly well in Fig. 1c, provides a way to estimate effects of allele substitutions. Among BXD strains the substitution of both B alleles with D alleles at Nnc1 is associated with an increase of approximately 10,000 ganglion cells. The intermediate population size of F1 progeny from strains with high and low cell numbers indicates that the mode of gene action is largely additive (1).
Variation in neuron number is often correlated positively with variation in brain weight (15, (16). The correlation across the BXD and parental strains is +0.54, a highly significant value (r2 = 0.29, F [1,24] = 10.0, P = 0.004). The polarity of phenotypes of the parental strains is reversed compared to that for ganglion cell number: C57BL/6J has lower mean cell number but has a substantially heavier brain than DBA/2J: 476 ±; 3 mg versus 392 ±; 5 mg. However, alleles at marker loci close to Nnc1 do not correlate well with brain weight (r2 = 0.13 at Tstap91A). Furthermore, in the BXH set, the correlation between brain weight and neuron number is negative (r = -0.3). This indicates that QTLs controlling variation in retinal ganglion cell number do not have notable effects on brain weight and therefore do not have global effects on neuron number in the CNS. However, given the large number of distinct cell populations in the CNS, Nnc1 may well have pleiotropic effects on other CNS populations.
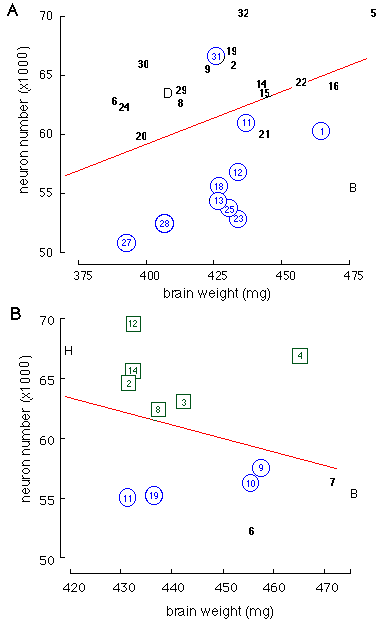
Regression analysis of brain weight on retinal ganglion cell number in RI strains. In both scattergrams the numbers in scatterplots correspond to particular strains listed in Tables 1 and 2. A, Scattergram of the BXD strains. The C57BL/6J parental strain is labeled with a B, and the DBA/2J parental strain is labeled with a D. The strains that are circled have the B type allele at the Tstap91A locus on Chr 11. Only strain BXD31 is discordant, with a B allele but a high cell population. Brain weights have been corrected for differences in sex, age, and body weight. The equation for the regression line is y = 23.5 + 0.09x, where y equals neurons (x1000) and x equals brain weight in milligrams. B, Scattergram of the BXH strains. The C3H/HeJ parental strain is represented by the letter H. Strains that are circled have the B allele at the Scya3 locus on Chr 11. (Tstap91A has not been mapped on the BXH strains, but Scya3 is a locus known to map 5-10 cM proximal to Tstap91A.) Strains that are boxed have the B allele at the Ssdh1 locus on Chr 4. The equation for the regression line in B is y = 114 - 0.12x.
We have begun to assess the the specificity of Nnc1 within the eye and retina (17). Variation in the size of the ganglion cell population does correlate positively with eye weight in BXD strains (r = 0.55) and retinal area (r = 0.52). However, as is true for brain weight, there is no signficant correlation between eye weight and alleles at loci on mid-distal Chr 11. To determine whether Nnc1 affects other cell populations in retina, we counted cells within the inner and outer plexiform layers of C57BL/6J, DBA/2J, BXD9, BXD13, BXD22, BXD23, BXD28, and BXD32 mice. There are large differences between cases and strains—from a low of 6.2 ± 0.3 cells per radial column in the photoreceptor layer in a BXD32 case with a ganglion cell population of 85,600, to 11.5 ± 0.5 cells in a BXD28 case with a ganglion cell population of 43,600. The correlation coefficient between ganglion cell number and the cell depth of the photoreceptor layer is -0.32 (95%; confidence interval of r is -0.71 to +0.21). However, the correlation coefficient between numbers of ganglion cells and cells in the inner nuclear layer (amacrine, bipolar, horizontal cells, and Muller glial cells) is +0.53 (confidence interval from +0.05 to +0.81). Collectively, these correlations suggest that Nnc1 may have effects on cell number in the inner nuclear layer but does not have effects on photoreceptor number.
We have mapped a major QTL that has a remarkably large effect on the size of the retinal ganglion cell population in mice. Replacing both C57BL/6J alleles with DBA/2J alleles at the Nnc1 locus generates a difference of about 10,000 cells—a 20%; increase in neuron number. The large effect has allowed us to map this QTL using a modest number of recombinant inbred strains. One of the principal advantages of recombinant inbred strains for this type of quantitative analysis is the ability to reduce non-genetic sources of variance by repeatedly phenotyping the same genotype. A second key advantage is that the genetic variance among a set of RI strains is 2- to 4-fold greater than that among corresponding intercross or backcross progeny (note 2). This improves the power of a search for QTLs.
Several investigators (18-20) have suggested that RI strains be used primarily to highlight chromosomal intervals that might be worth subsequent analysis using larger panels of backcross and intercross progeny. We suspect, however, that QTLs with sufficiently large effects may prove to be more common than expected when narrowly defined morphological traits are targeted for analysis (18, 19, 21). An alternative strategy that may prove productive is to increase the number of RI lines included in an analysis and to pool across partially or completely independent RI sets. In our case, the first 12 BXD strains that we studied highlighted several candidate intervals, including mid-distal Chr 11. The addition of the remaining 14 BXD strains winnowed the initial list of candidates and greatly strengthened linkage near Tstap91A. Adding the 12 BXH recombinant strains enabled us to detect secondary QTLs that we would otherwise have missed. By controlling for the effects of QTLs detected initially in one or the other set of RI strains, we were able to greatly improve the strength of linkage to the Nnc1 locus. This bootstrap procedure may be particularly useful for mapping quantitative traits already known to differ substantially among the 10 to 20 strains from which recombinant inbreds have been generated.
Nnc1 maps between Hoxb and Krt1 (12). This region (22) contains three excellent candidates for Nnc1—Rara, Thra, and Erbb2. All three genes encode receptors known to be expressed in retina early in development. It is also known that changing the concentrations of the ligands of these receptors—retinoic acid, thyroxine, and neuregulin—affects the proliferation and survival of retinal cells (23-30). For example, an increase in thyroxine triggers the production of new retinal ganglion cells that specifically have uncrossed projections in Xenopus (31). Retinoic acid has also been found to have effects on retinal cells. For example, the addition of exogenous retinoic acid increases rod production at the expense of amacrine cells (29). Finally, neuregulin, a ligand that activates the erbB2 tyrosine kinase receptor (32), promotes ganglion cell survival in culture (24). To assess the developmental mechanisms generating variation in retinal ganglion cells among strains we have counted these cells at birth, before the onset of naturally occurring cell death. Our results suggests that Nnc1 affects ganglion cell production rather than ganglion cell death (33).
A growing number of loci are already known to influence numbers and ratios of retinal cell types when mutated, knocked out, or overexpressed. The list includes pearl (34), Brn3 (35), Pax6 (36), Mitf (37, 38), Chx10 (39) Hes1 (40), Bst (41), Notch1 (42), Ccnd1 (43), Bdnf (44), Fgf (45), Ngf (46), and Bcl2 (47-49). The loss of Brn3b, for example, reduces ganglion cell numbers by 60 to 70%. Brn-3b has been shown to ge down-regulated by retinoic acid (58). In contrast, overexpression of Bcl2 attenuates normal cell death, allowing twice the normal number of ganglion cells to survive. It is possible that normal alleles at these loci have more subtle effects and could account for some of the normal genetic variance not produced by alleles at Nnc1. Surprisingly, loss of the alpha-1 isoform of the retinoic acid alpha receptor, one of the candidates for Nnc1, has no known effect on the eye or retina (50). Mutant and null alleles at this and other retinoic acid receptors may have more subtle quantitative effects, a possibility that we are now testing. The fact that so many null mutants are viable and apparently normal has led to the idea that key developmental mechanisms are often controlled by the products of several closely related genes. Some of these redundant loci may function primarily as QTLs and maintain a reservoir of allelic variants.
The remarkable speed of brain evolution in response to shifts in selective pressure (4, 15, 51) is dependent upon allelic variants at loci that control the size of neuron populations by proliferation and cell death (4, 15). The four-fold increase in the size of the cerebellar cortex (52) that has occurred over the past several million years in the lineage leading to modern humans was probably brought about by gene modifications that have increased proliferation in select groups of rhombencephalic progenitor cells. The rapid reduction in neuron number in the cat's retina and dorsal lateral geniculate nucleus over a period of less than 20,000 years was probably brought about by changes in severity of natural cell death (15). This study has demonstrated that a focused approach that exploits normal variation can uncover polymorphic loci with large and specific effects on CNS cell populations. We anticipate that rapid progress in mapping QTLs with prominent effects on CNS traits will lead to a better understanding of the basis of normal variation in CNS structure and function, and ultimately, will lead to a better understanding of the genetic basis of brain evolution.
Acknowledgments. We thank K. Troughton, R. Cushing, and T. Hurt for technical support. We thank Dr. K. Manly for his program, Map Manager QT, and Drs. B. Taylor, J. Cheverud, and R. Elliott, and D. Rice for comments on drafts. This work was supported in part by grants from the National Institute of Neurological Disease and Strokes (to R.W), and the National Eye Institute (to R.W. and D.G.).
| Table 1. Cell number and brain weight in BXD strains@ | |||||
|---|---|---|---|---|---|
| Strain | Mean (x1000) | SE# (x1000) | Type** | Brain* (mg) | Residuals+ (cells x 1000) |
| C57BL/6J | 55.4 | ± 0.8 | L | 475 | . |
| DBA/2J | 63.4 | ± 1.2 | H | 412 | . |
| BXD1 | 60.3 | ± 1.1 | I | 465 | -4.64 |
| BXD2 | 65.9 | ± 1.8 | H | 432 | 3.88 |
| BXD5 | 75.5 | ± 1.3 | H | 526 | 5.06 |
| BXD6 | 62.7 | ± 0.9 | H | 388 | 4.74 |
| BXD8 | 62.8 | ± 2.1 | H | 412 | 2.72 |
| BXD9 | 65.6 | ± 1.7 | H | 422 | 4.56 |
| BXD11 | 61.0 | ± 1.1 | I | 437 | -1.37 |
| BXD12 | 56.8 | ± 2.1 | L | 434 | -5.39 |
| BXD13 | 54.7 | ± 1.7 | L | 427 | -6.76 |
| BXD14 | 64.0 | ± 1.6 | H | 442 | 1.21 |
| BXD15 | 63.8 | ± 1.1 | H | 443 | 0.86 |
| BXD16 | 64.0 | ± 1.3 | H | 469 | -1.29 |
| BXD18 | 55.1 | ± 0.9 | L | 427 | -6.44 |
| BXD19 | 67.1 | ± 1.3 | H | 431 | 5.14 |
| BXD20 | 59.9 | ± 1.9 | I | 398 | 1.00 |
| BXD21 | 60.0 | ± 1.5 | I | 443 | -2.97 |
| BXD22 | 64.5 | ± 1.1 | H | 457 | 0.22 |
| BXD23 | 53.0 | ± 1.0 | L | 434 | -9.13 |
| BXD24 | 62.4 | ± 1.0 | H | 391 | 4.11 |
| BXD25 | 53.8 | ± 1.5 | L | 431 | -8.02 |
| BXD27 | 50.8 | ± 1,1 | L | 393 | -7.62 |
| BXD28 | 52.4 | ± 1.9 | L | 407 | -7.31 |
| BXD29 | 63.6 | ± 1.4 | H | 413 | 3.31 |
| BXD30 | 66.0 | ± 1.3 | H | 399 | 7.05 |
| BXD31 | 66.6 | ± 1.2 | H | 426 | 5.09 |
| BXD32 | 75.8 | ± 2.2 | H | 434 | 13.59 |
| Table 2. Cell number and brain weight in BXH strains@ | |||||
|---|---|---|---|---|---|
| Strain | Mean (x1000) | SE (x1000) | Type | Brain@ (mg) | Residuals |
| C57BL/6J | 55.4 | ± 0.8 | L | 475 | . |
| C3H/* | 67.0 | ± 1.7 | H | 427 | . |
| BXH2 | 64.6 | ± 2.2 | H | 431 | 2.46 |
| BXH3 | 63.0 | ± 2.1 | I | 442 | 2.18 |
| BXH4 | 66.9 | ± 3.3 | H | 465 | 8.88 |
| BXH6 | 52.3 | ± 1.4 | L | 455 | -6.90 |
| BXH7 | 56.5 | ± 2.2 | I | 471 | -0.85 |
| BXH8 | 62.4 | ± 2.2 | I | 437 | 1.01 |
| BXH9 | 57.5 | ± 1.8 | I | 457 | -1.54 |
| BXH10 | 56.2 | ± 1.0 | I | 455 | -3.03 |
| BXH11 | 55.0 | ± 1.7 | L | 431 | -7.10 |
| BXH12 | 69.5 | ± 1.0 | H | 432 | 7.50 |
| BXH14 | 65.6 | ± 2.5 | H | 432 | 3.56 |
| BXH19 | 55.3 | ± 2.8 | L | 436 | -6.17 |
| Table 3: Strain distribution patterns of retinal ganglion cell number and loci on Chr 11 | ||
|---|---|---|
| Locus | cM* | 1 2 5 6 8 9 1 2 3 4 5 6 8 9 0 1 2 3 4 5 7 8 9 0 1 2 |
| RGC Number | ||
| D11Ncvs58 | 56 | |
| Hoxb | 56 | |
| Tstap91A | 57 | |
| Krt1 | 58 | |
| Mpmv8 | 62 | |
