




 |
 |
 |
 |
 |
|
|
|
|
|
||
|
EXPERIMENTAL PLAN |
|||
|
Principal Investigator/Program Director Williams,Robert W. |
|||
New methods and their advantagesAim 1: Construction of mouse 3D brain atlases and navigation softwareIn Aim 1, we propose to construct eight 10-mm isotropic brain atlases: two stains (Nissl and fiber), two strains (C57BL/6J and DBA/2J), and both sexes. The brains will be manually delineated. A transformation function for mapping coordinates across the brains will be established. To view the atlases over the Internet, a JAVA version of MacOStat, our existing atlas viewer, will be developed and released. 3D atlas reconstructionThe sections to be aligned in the atlas construction phase will be imaged at a resolution of 4.5 mm. Recently, we have begun examining whether the volumes could be reconstructed without the aid of blockface imaging. Instead we have performed section-to-section alignment using AIR (Woods et al. 1998 a, b). This method yields surprisingly good global alignment (Fig. 2), and we have used it to generate a 30-mm isotropic reconstructed brain from celloidin-embedded sections (Fig. 3). Because rigid-body transformation was used for this alignment, cutting-induced distortion reduced reconstruction accuracy. To examine whether nonlinear registration will be required to reconstruct 10 mm isotropicatlases, we aligned consecutive sections using punctate fiducials (blood vessels). The residual misregistration was found to be about 4550 mm. To overcome this reconstruction error, fiducials will be used to drive nonlinear transformation. After rigid-body inter-section alignment, distortion will be corrected by selecting ROIs in one section, cross-correlating these with nearby regions on adjacent sections, and obtaining displacement vectors. We hypothesize that overlap of cells that occur on both sections will cause a correlation peak. If the two sections contain information from linear structures (e.g., blood vessels) or planar structures (e g., cell layers or ventricle edges) that are oblique relative to the section plane, then the correlation peaks due to these features may be displaced. The possibility of such bias will be mitigated by (1) avoiding regions that contain such structures and (2) high-pass filtering the two images to suppress broad correlation peaks. The number of such alignment points needed depends on the spatial frequency content of the distortion field, which will be established empirically by constructing such a field from many windows on a sample of section pairs. Preliminary data suggest that about 25 (5 x 5) masks, each 200 x 200 mm, will suffice. A distortion field from section i to section I+1 will be constructed by assuming that the I th section is distortion-free. Spline interpolation will be used. After all such maps are collected, they will be globally relaxed by minimizing the total mechanical distortion (strain) energy over the brain. Our overall objective is to achieve an unprecedented alignment accuracy of better than 10 mm.
Atlas delineationDr. Nissanov, Dr. Williams and Ms. Bertrand, all of whom have expertise in neuroanatomy, will delineate the atlases in three dimensions using available software. For this phase of atlas creation, the anisotropic volume will be employed (4.5 mm in plane, 10 mm in the orthogonal axis). To accomplish this task, we have built a software tool, BRAIN (available from the Computer Vision Center for Vertebrate Brain Mapping at Drexel), and have employed it in delineating our 2D-atlas fully (Nissanov and Bertrand 1998a) and the 3D-atlas partially (Nissanov et al., submitted). Delineation will be smoothed in 3D using morphological operators. In addition, other investigators will have the option of inserting their own delineations. Thus, users can select which anatomical template they wish to employ. Complete delineation of each atlas (approximately 600 VOIs per side, 1200 total) will take approximately 6 months. They will be made available as they are generated. Cross atlas alignment
Our
objective is to define a single standard coordinate system for the MBL.
One atlas will be selected for that purpose and the others will be
transformed into that system. Point-to-point mapping will first be
determined by an affine transformation computed with our previously
developed 3D distance-based alignment algorithm (3DBA), followed by a
small radial spline transformation. The transformations will be
computed with a mixed fiducial point and fiducial surface alignment
algorithm based on the method in Gabrani and Tretiak (1999). The fiducials
will consist of a set of about
100 three-dimensional points delineated manually. These and the outer
surface will be used to drive the algorithm. The required software is
already available at Drexel.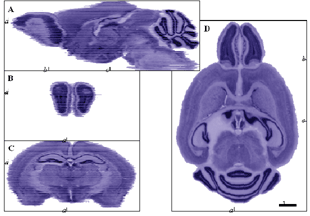 Figure 3. A 30 m isotropic 3D reconstruction of C57BL/6J brain. Images celloidin sections were aligned using AIR (rigid-body). The original cutting plane was horizontal (far right, D). Sagittal (top, A) and two coronal planes (lower left, C and D) were obtained by resampling the volume. Location of views is cross-indicated by tick marks with adjacent notation referring to the panel of the given view. No gray value alignment (e.g., Nissanov et al., submitted) was performed. Scale bar applies to all panels. NeuroTerrain versions 1.0 and 2.0MacOStat, atlas navigation software for the Macintosh platform, is available from the Brain Mapping Center at Drexel. This application allows users to interactively define an arbitrary plane of view through a 3D data set. The view is dynamically updated as the user rotates or translates about x, y, or z axis. The atlas also supports VOI display. The outline generated by intersection of VOIs and the plane of view can be superimposed over the atlas. MacOStat currently is written in C++ and runs on the Macintosh operating system. We will port it to JAVA to support navigation over the Internet (NeuroTerrain) and will work with Dr. Rosen to achieve a smooth interface with the MBL. A consideration is the speed of navigation over the Internet. A view through the 10-mm isotropic atlas planes along the greatest dimension requires 800 KB of computer storage space. In MacOStat we have achieved dramatic reduction (44%) in memory (both disk and RAM) footprint using a macrovoxel data representation (Nissanov et al., submitted). Voxel data are organized first into macro voxels, which taken together form the entire volume. An internal coordinate system relative to the macro voxels is used for slicing operations. The whole number portion of a coordinate triple specifies the macro voxel containing that point, and the fractional portion of the coordinate triple specifies the micro voxel within the macro voxel. Any macro voxel in which all micro voxels have the same value is eliminated. In practice, any macro voxel that is all white (the image background) is eliminated because the class representing the volume map will return a white voxel for any missing macro voxel. Thus, in navigating through the atlas, the data transfer requirement is reduced to less than 450 KB per view. With current networks, this translates to about a view every 5.5 sec. To accelerate this process, we will employ a progressive encoding scheme that supports a 10-fold speed increase during coarse navigation. We estimate release of the first navigator version will be 9 months after commencement of the grant period. A manual and a tutorial will be distributed online with the applet at no charge.MacOStat currently supports simultaneous viewing of homologous arbitrary planes through multiple volumes. This feature will be incorporated into the NeuroTerrain version (v 2.0) of the navigator by the end of Year 02. |
|||
|
Next Topic |
|||
|
|
|||