|
| ||||||
|
| ||||||||
|
| ||||||||
Home  Publications Publications |
|
|
Genetic Analysis of Variation in Neuron Number Richelle Cutler Strom Chapter 4: Genetic and Environmental Control of Retinal Ganglion Cell Variation Introduction Evidence of global genetic control over cell number is found from the linear scaling of brain structures with brain size across species (Finlay and Darlington, 1995). There is also evidence of localized genetic control over cell number within particular brain structures. The genetic control within brain structures is necesssary for the conservation of neuron number in the event of species-specific adaptations. Some examples of species-specific adaptations associated with neuron number are the capacity for bird song and foraging memory in birds, and the lateralization of language in humans. With each species adaptation, selective pressures for behavior must have acted upon the natural variation in the neuron population within the species. Examining the genetic bases of variation in discrete cell population within a species will shed light on the genetics of species-specific adaptations in neuron number. In this study, I examine the genetic and environmental control of variation in a single well-defined neuron population, the retinal ganglion cell population, among different inbred strains of mice. Retinal ganglion cells are the sole projection neurons relaying visual information from the eye to the brain via their axons in the optic nerve. The morphological and physiological attributes of retinal ganglion cells have been described in many different species (Rodieck and Brening, 1983). The relative simplicity of the retinal circuitry and its accessibility make the retina an attractive model system for studying CNS development. Consequently, a lot is known about the development of the retina, and in particular ganglion cell differentiation. The differentiation of a large portion of ganglion cell has been shown to require a POU-domain transcription factor, Brn-3b (Gan, 1996). Other factors that may be involved in ganglion cell differentiation are fibroblast growth factor (Guillemot and Cepko, 1992) and the transcription factor RPF-1 (Zhou et al., 1996). Estimating numbers of distinct neuron populations can be tedious and error prone. However, retinal ganglion cell number can be estimated precisely and quickly by estimating the number of axons in the optic nerve. This is important because the precision of heritability estimates and gene mapping rely on the ability to obtain accurate phenotype data. In order to map genes that control ganglion cell number, specifically, the variance attributed to cofactors, such as brain weight, body weight, sex, and age, needs to be minimized. In this chapter, I estimate the variance in ganglion cell number associated with cofactors using linear regression. I also examine the precision with which ganglion cell numbers can be estimated. Finally, as a prelude to mapping QTLs responsible for variation in ganglion cell number between mouse strains I determine the heritability of ganglion cell number and the minimum number of effective genes.
Materials and methods Mice Optic nerves and brains were taken from mice, belonging to18 standard inbred strains. All of these strains were obtained from the Jackson Laboratory (Bar Harbor, ME). Optic nerves and brains were taken from 26 BXD recombinant inbred strains and two groups of intercross progeny generated from crosses between BALB/cJ and CAST/Ei (CCASF2) and between BXD32 and CAST/Ei (32CASF2). I also obtained optic nerves and brains from five isogenic F1 hybrids, (32CASTF1, CCASF1, CAF1/J, B6AxCF1/J, PLSJF1). The hybrids CAF1/J and PLSJF1 were obtained from the Jackson Laboratory and the remaining were generated in the University of Tennessee animal colony. Data for the F1 hybrids and their parents can be found in the Table 4.3. In three of the five hybrids, the parents have large differences in ganglion cell number. For example, 32CASF1 progeny were generated from parents BXD32 and CAST/Ei. The BXD32 strain has the highest ganglion cell number measured among the strains (75,800 ± 1,980) and CAST/Ei has the lowest ganglion cell number (45,000 ± 1,000). Some of the strains studied carried mutations affecting the retina. Six strains carried a mutation in the ß -phosphodiesterase gene (C3H/HeJ, C3H/HeSnJ, CD-1, PL/J, SJL/J, and MOLD/Rk). A mutation in the ß -phosphodiesterase gene leads to retinal degeneration (rd). Retinal degeneration in the most extreme cases results in the complete loss of rod photoreceptors by 2 months of age. However, it is not evident that rd affects ganglion cell number, since all the strains carrying rd, with the exception of MOLD/Rk, have average ganglion numbers and one strain even has a high ganglion cell number. The following strains carry the albino allele of the gene tyrosinase (A/J, AKR/J, BALB/cJ, CD-1, 129/J, 129/SvJ, NZW/LacJ, SJL/J). Albino mice typically have a reduction in the proportion of retinal ganglion cells with uncrossed projections, but there is no association between the albino allele and adult ganglion cellnumber (Rice et al., 1995). Tissue Fixation Optic nerves were dissected from mice transcardially perfused with EM grade fixative. The optic nerves were processed in 96 well microtiter plates. The procedure involved gradually dehydrating the nerves with increasing concentrations of alcohol, staining with 2% osmium, and infiltration with Spurr’s resin. Embedded nerves were thin-sectioned, placed on formvar-coated grids, and stained with lead citrate and uranyl acetate. Estimating ganglion cell number Ganglion cell number was estimated by counting their axons within the optic nerve. Previous studies have demonstrated that axon counts are reliable estimates of ganglion cell number, (Chalupa et al., 1984; Lia et al., 1986; Perry et al., 1983; Rice et al., 1995; Williams et al., 1986), see table 2). Nerves were photographed in a grid pattern on a JEOL EX2000II electron microscope typically at a magnification of ~ x12,000. The nerve area was determined by photographing the entire nerve at low magnification, usually at 200x or 250x. Immediately after sampling the nerve at high magnification and photographing the entire nerve at low magnification, the high and low magnifications were calibrated by photographing a grid replica (2160 lines/mm, obtained from EMS, Fort Washington, PA). Axons were counted within a 63 x 86 mm counting frame drawn on the micrograph. To give unbiased estimates only those axons intersecting the top and right edge of the frame were included in the count. Total axon estimates were calculated by multiplying the mean axon density (per micrograph area) by the total optic nerve area.
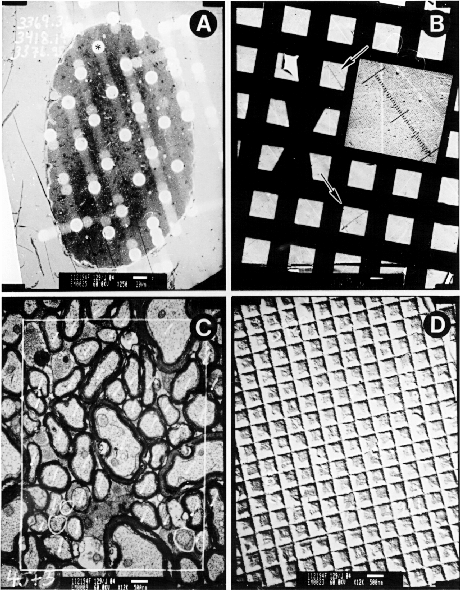
Figure 4.1. Method for estimating ganglion cell numbers. Ganglion cell number for strains, species, and subspecies of mice. The major subdivisions of mice are indicated in the left margin and are described in the Materials and Methods. Numbers for each strain denote average ganglion cell number, standard error of the mean (SEM) and the number of cases. These four prints were made from negatives used to estimate ganglion cell number. A and B are low-power electron micrographs of an optic nerve cross-section (A) and calibration grid (B). C and D high-power electron micrographs of an optic nerve sample site (C) and calibration grid (D). The white spots on the nerve in A are areas that were bleached by the electron beam during sampling at high-power. The asterisk in A marks the site that is shown at high magnification in C. The axon margin was outlined with a sharpie marker using the aid of a light microscope. The area of the nerve was calculated by tracing the outlined nerve on a digitizing tablet. The two arrows in B mark the sites where delimits spanning 80 grid units was etched into the negative with a small scalpel and this distance measured under a dissecting microscope with a digital caliper. Axons were counted within a counting frame drawn in C. The axon count in this sample is 45 myelinated and 3 unmyelinated fibers (circled). The area of the counting frame was calibrated using calibration grating in D. The dimensions of the calibration grid units are 0.463 x 0.463.
Heritability The broad-sense heritability of ganglion cell number was calculated from the ratio of the genetic variance (VG) over the total phenotypic variance (VP) found in the outbred strains. VG was calculated by subtracting the average variance in ganglion cell number, found within the two outbred strains (CD-1 and CARL/ChGo), by the average environmental variance found within the inbred strains. The environmental variance was obtained by averaging the variance within 18 inbred strains and 6 F1 hybrids. The average variance served to reduce the effects of variance deviation resulting from unique genotype–environment interactions. A close approximation to heritability in the narrow-sense was made using the Hegmann and Posidente (1981) method. The number of effective loci influencing ganglion cell number in the F2 intercross mice was calculated using the Castle-Wright equation.
Results Variation among strains, species and subspecies The ganglion cell number among standard inbred strains of mice range from 50,600 in A/J to 69,000 in BXD32 (Table 4.1). The average ganglion cell number for the 18 standard inbred strains is 59,300 ± 1130 (n = 150). The average coefficient of variation (CV) for inbred strains is 6.9%. The variance among strains is much greater than within strains (F(16,119) =15.0, p < 0.0001). There are large differences in ganglion cell number between closely related inbred strains. For example, the strains 129/J and LP/J originated from a common ancestor in the mid-1920s, but their average ganglion cell population differs by ~17,000. A difference of 11,000 ganglion cells was discovered between shipments of C57BL/6J mice from the Jackson Laboratory. The different shipments of C57BL/6J originated from different breeding rooms at the Jackson Laboratory. No non-genetic factors were identified that could explain the differences in ganglion cell number of the mice residing in the two rooms. Thus, the difference in ganglion cell number is possibly due to a new genetic mutation arising in the separate breeding colony.
Table 4.1. Average ganglion cell number for 18 standard inbred strains
*Pooled data from pigmented and coisogenic albino mice. The statistics SD, SE, and CV were corrected for bias because of small sample size ( Sokal and Rohlf, 1981). Data for the strain 129/SvJ is new and is not published in Williams et al. (1996).
Ganglion cell number among species and subspecies ranges from 45,000 to 64,300. The average CV for these strains is 7.4% (Table 4.2). Note that the strain CARL/RpGo is outbred and as expected has a higher degree of variation, CV = 14%.
Table 4.2. Average ganglion cell number for inbred representatives of wild strains
*MOLD/Rk carries the rd allele and there is a high incidence of necrotic axons in the optic nerve of older mice. Estimates from the two youngest MOLD/Rk cases (49,200 and 47,200) are probably better representatives. **CARL/ChGo is an outcrossed strain.
The ganglion cell probability distribution of the 57 inbred strains, including 19 inbred laboratory strains and 38 recombinant inbred strains (Fig 4.2). The probability distribution has three modes at 55,500, 63,500, and 75,500 and indicates that there are three distinct phenotypes. The third mode consists of the strains BXD5 and BXD32, which have exceptionally high ganglion cell numbers that are 20% above their high parental strain—DBA/2J. A Kolmogorov–Smirnov goodness-of-fit test confirms that the inbred strain distribution is not normally distributed (D = 0.245, p < 0.01)
Figure 4.2. Probability density of retinal ganglion cell number for 57 inbred strains. The large filled-in function is a summation of individual normalized probability densities of the ganglion cell averages from 57 strains. The two small filled-in functions labeled B and D are examples of the individual probability densities computed from C57BL/6J (B) and DBA/2J (D) strain averages. The black line is the expected Guassian distribution based on the mean of all strains (61,730 ± 6,310).
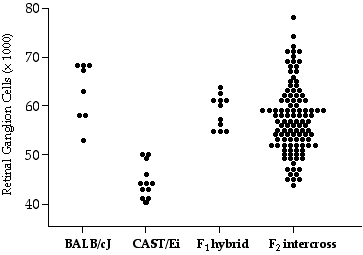
The variance in ganglion cell number within the heterogeneous crosses is higher than the variance within the inbred strains (Table 4.3). The average of corrected ganglion cell number in the CCAS F2 progeny is 56,000 ± 6000 (SD). The distribution of raw ganglion cell numbers in the CCASF2 is broad, ranging from a low of 42,000 to a high of 70,000 (Fig 4.3). The broad range in ganglion cell numbers indicates that multiple genetic factors have segregated that can modify ganglion cell number. The number of genes modulating ganglion cell number was estimated to be one in CCASF2 cross and three in the 32CASF2 cross.
Table 4.3 Average ganglion cell population in heterogenous mice.
*CD1/Go is an outbred laboratory strain.
Figure 4.3. Scatterplot of retinal ganglion cell number for CCASF2, CCASF1, and parental strains.
Technical variation Reliability was estimated by sampling a set of 69 nerves two to four times. Usually an adjacent nerve section was photographed and this was done months to several years after the first count. The average of the first count was 58,967 ± 1145, whereas that of the second count was 58,667 ± 1,361. The test-retest correlation coefficient was 0.83. The mean difference between pairs of estimates was 4,080, and the average SD of sets of replicates was 3,116. The technical coefficient of variation between repeated samples of ~25 micrographs averaged 6.3 ± 0.5% SE. I calculated the sampling variance across repeated samples and estimated that 60–70% of the technical variance is associated with sampling error. Variance in calibration errors and axon discrimination can account for the remaining variance. The coefficient of variation for the standard inbred strain averaged 6.9%. Thus, after taking into consideration the technical coefficient of variation the corrected environmental coefficient of variation is 2.8%. Correlation between neuron number, ages, sex, body weight, and brain weight The effects of age, sex, body weight and brain weight on ganglion cell number within–strains were examined by calculating within strain z-scores for retinal ganglion cell number and comparing this value to age, sex, body and brain across strains. The correlation between age, sex, body weight, brain weight and within–strain ganglion cell number within the 18 standard inbred strains were approximately zero, (n = 124). In these strains, the sex ratio was 50:50 and ages ranged from 30 to 200 days. The correlation between brain weight and ganglion cell number across strains is +0.38, which is highly significant, (t = 6.54, n = 184, p < 0.001). The correlation between brain weight strain averages and ganglion cell strain averages is +0.46, which is also significant at p < 0.02. The corresponding coefficient of determination (r2) is 21.5 %. The correlation between ganglion cell numbers and brain weights regressed with respect to sex, age, and body weight is roughly the same, r = 0.42. Within BXD recombinant strains, ganglion cell number was not correlated with brain weight, body weight, sex, or age. However, across strains variation in ganglion cell number was significantly correlated with brain weight, r = 0.42. The correlation coefficient was reduced to 0.36 when brain weight was corrected for variation in sex, age, and body weight. In the CCASF2 cross variation in ganglion cell number was significantly correlated with brain weight explaining 26.8% of the variation in ganglion cell number (p = 0.0001). Body weight was also significantly correlated with ganglion cell number (r2 = 11.5%, n = 92, p = 0.001), a product of the high correlation between body and brain in CCASF2. However, adding brain weight to the regression with body weight abolishes any correlation with body weight. There was no correlation between ganglion cell number and sex, age or parity in the CCASF2 cross. To reduce the effect of brain weight variation on ganglion cell number, the ganglion cell numbers of CCASF2 mice were standardized to that of a 437 mg brain, the average brain weight of the F2 progeny. Correcting for differences in brain weight resulted in a 25% reduction in variance of ganglion cell number among CCASF2 progeny. Variation in ganglion cell number was also significantly associated with brain weight in the 32CASTF2 cross, but explained only 15.9% of the variation (p < 0.0001). There was also no correlation between ganglion cell number and sex, age or parity in the 32CASF2 cross. Heritability and gene dominance Broad-sense heritability calculated in outbred strains CD-1 and CARL/ChGo is 0.73 and 0.68, respectively. Narrow-sense heritability of retinal ganglion cell number in the standard inbred mouse strains is 0.48. Narrow-sense heritability of ganglion cell number in the BXD strains is 0.47. Heritability for ganglion cell number in the CCASF2 cross is 0.90 and was reduced to 0.80 with corrected numbers. Heritability of ganglion cell number in the 32CASTF2 cross is 0.50. Estimates of the effective gene number modulating ganglion cell number are one in the CCASF2 cross, 2.5 in the 32CASF2 cross, and 2.4 in the BXD strains. Due to the required assumptions for this calculation, these estimates are likely to be underestimates of the true gene number. Three hybrids, CCASF1, 32CASF1, and PLSJF1/J, exhibit dominance for high ganglion cell number alleles (Table 4.4). For example, the average ganglion cell number for CCAS F1 hybrids is 59,500 ± 3,400 (SD) and is roughly 5,000 cells higher than the parental midpoint. The CV of variation within the hybrids in Table 4.4 averages 5.7%. This CV average is not significantly different from the CV of 6.9% found in the inbred strains (one tailed t-test, p = 0.15). The heterozygous state in some hybrids increases fitness that provides a buffer against developmental and environmental effects. However, there is no evidence that the ganglion cell number in these hybrids is more stable than in the homozygous strains.
Table 4.4. Average ganglion cell number in F1 hybrids and their parental strains
Discussion The significantly lower variance in ganglion cell number within strains compared to across strains demonstrates that genetic differences are involved in the variation in ganglion cell number among strains. Heritability estimates for variation in ganglion cell number are high, ranging from 0.80 in CCASF2 to 0.47 in the BXD strains. Estimates of heritability within crosses generated from parental strains that are widely divergent, such as BALB/cJ, and CAST/Ei parentals for CCASF2, are typically inflated (Lynch and Lynch, 1992). In addition, the estimates of broad sense heritability in the intercrosses include the effects of gene dominance effects and epistatic interactions among loci are in the neighborhood of 80%. Nevertheless, these estimates are near the 80% estimate of genetic control over granule cell number in the dentate gyrus of mice (Wimer and Wimer, 1989). Heritability estimates such as these are based on ratios between genetic and nongenetic variance. Consequently, minimizing environmental variance increases measured heritability. In this study, all mice were reared in a pathogen-free laboratory environmentÑa situation that eliminates numerous sources of environmental differences, and that almost certainly increases estimates of genetic control compared to dispersed wild populations of mice. The ganglion cell numbers of CCASF1 and 32CASF1 exhibit dominance with respect to the high parental. This dominance could result from hybrid vigor (see discussion in Chapter 2). In addition, the dams that rear the F1 hybrids (BALB/cJ and BXD32) have a larger body size compared to that of CAST/Ei. The BALB/J and BXD32 neonates are larger in size compared to CAST/Ei neonates. Case in point, when reciprocal matings were performed, the CAST/Ei females were dieing in labor trying to birth these larger size hybrid neonates untypical of their species.The genetic factors producing a small body size in CAST/Ei could have pleiotropic effects on brain weight and neuron numbers. Without the body size restriction imposed, the CAST/Ei contribution the CCASF1 ganglion cell number might raise above the midparental range. The dominance of the CCASF1 is also reflected in the distribution of the CCASF2 (Fig. 4.3). An analysis of isogenic animals makes it possible to assess the consistency with which the genome guides the generation of traits such as neuron number. When environmental differences are minimized, the residual variance is due to microenvironmental effects and developmental noise. In a systematic analysis of the grasshopper nervous system a remarkable level of variation in neuron number was found both within and between isogenic groups that had been reared in a tightly controlled environment. In one exceptional group, half of the animals had the standard set of six ocellar interneurons whereas the other half had seven to nine interneurons (Goodman, 1979). In contrast, in a survey of optic nerves from a set of more than 100 isogenic crustaceans (Macagno, 1980) found only a single exception to the rule of 176 axons. Thus, the developing nervous system is capable of of excruciating control. However, it is not known how stringently large populations are developmentally regulated in the vertebrate CNS. The average corrected coefficient of variation in ganglion cell number is 2.8%. Thus, the developmental control of ganglion cell number is stringent. Although, coefficients of variation do range from 2.0% in NZW/LacJ to 11% in C57L/J indicating that the degree of developmental stringency is under genetic control. In addition, these differences could result from some of sampling artifacts discussed in the methods Variation in ganglion cell number was significantly correlated with brain weight across strains and within the intercrosses. However, there was no correlation between within strain z-scores and brain weight. The absense of a correlation between ganglion cell number and brain weight within strains suggests that the significant correlation found across strains is not due to common non-genetic factors, such as the maternal environment. Thus, the association of ganglion cell number and brain weight is due to the global variation in cell number that originates early during development before the divergence of specific cell lineages. Furthermore, the variation in brain weight is a summation of the correlated variation of individual populations as distinct as the retinal ganglion cells. There was no decline in ganglion cell number with age in any of the mice studied. This finding is corroborated by a study reporting no age-related decline in ganglion number in the retina of young and old rhesus monkeys (Kim et al., 1996). There are many reports of age-related decrease of neurons in the cerebral cortex and cerebellum neuron populations, but there are also many reports of no age-related decrease in neuron number within distinct nuclei (Jacobson, 1991). In summary, a decline in neuron number is not an inevitable result of aging and may depend on the endogenous level of oxidative stress in a particular neuron type. The bimodality of strain averages is a surprising and important finding that provides evidence that there are single polymorphic genes that have comparatively large effects on neuron number. This conclusion is strengthened by the estimate from the Castle-Wright formula of one to three genes with large effect size modulating ganglion cell in the F2intercrosses. A speculation based on these findings is that variation in other neuron populations may also be controlled by relatively small numbers of quantitative trait loci that have major effects. The low gene number and high heritability of ganglion cell number suggests that it should be possible to map one or more genes with a quantitative effect on ganglion cell number.
|
Neurogenetics at University of Tennessee Health Science Center
| Text Only | Top of Page |
Mouse Brain Library | Related Sites | Complextrait.org