|
| |||||
|
| ||||||||
|
| ||||||||
Home  Publications Publications |
|
Quantitative Neurogenetics & QTL Mapping The BXD recombinant inbred strains are a versatile reference mapping panel for genetic dissection of complex phenotypes. These strains can be used in a relational fashion to combine multi-level data ranging from sequence gene expression analysis to brain and behavioral phenotypes. Published winter 2003 in Neuroinformatics by Elissa J. Chesler, Jintao Wang, Lu Lu, Yanhua Qu, Kenneth F. Manly and Rob Williams. The paper is available in PDF version. A comprehensive analysis of linkage maps of recombinant inbred strains with comparisons to F2, N2, and advanced intercross maps. We genotyped just over 100 RI strains using a set of approximately 1600 microsatellite markers. Published October 2001 in Genome Biology by Rob Williams, Jing Gu, Shuhua Qi, and Lu Lu. The papers is available both in PDF and HTML versions. Our own local HTML version includes updated links to new RI genotype data files. Users of RI strains (AXB-BXA, BXD, BXH, and CXB) will find the mapping data sets useful. Cancer susceptibility is a complex interaction of an individual's genetic composition and environmental exposures. Huge strides have been made in understanding cancer over the past 100 yr, from recognition of cancer as a genetic disease, to identification of specific carcinogens, isolation of oncogenes, and recognition of tumor suppressors. A tremendous amount of knowledge has accumulated about the etiology of cancer. Cancer genetics has played a significant role in these discoveries. Analysis of high-risk familial cancers has led to the discovery of new tumor suppressor genes and important cancer pathways. These families, however, represent only a small fraction of cancer in the general population. Most cancer is instead probably the result of an intricate interaction of polymorphic susceptibility genes with the sea of environmental exposures that humans experience. Although the central cadre of cancer genes is known, little is understood about the peripheral genes that likely comprise the polymorphic susceptibility loci. The challenge for cancer genetics is therefore to move forward from the Mendelian genetics of the rare familial cancer syndromes into the field of quantitative trait loci, susceptibility factors, and modifier genes. By identifying the genes that modulate an individual's susceptibility to cancer after an environmental exposure, researchers will be able to gain important insights into human biology, cancer prevention, and cancer treatment. Read more A short report of a symposium at the 2001 IBANGS meeting. An upbeat commentary in Behavior Genetics on complex trait analysis and "cloning" QTLs that we hope will counterbalance the gloomy assessment by Joe Nadeau and Wayne Frankel (2000). Our review (John Belknap and colleagues) is in a special issue of Behavior Genetics devoted to QTL mapping and complex trait analysis. Spread the word: QTL analysis is thriving, and yes they can be cloned. An introduction to mapping quantitative trait loci (QTLs) written for neuroscientists taking the 1998 Short Course in Quantitative Neuroanatomy. Updated August 2000 with new figures and text on RIX mapping. A review chapter from Mouse Brain Development that explains the power of QTL mapping in exploring CNS development. This review included original data on brain weight and neuron number in different strains of mice (C57BL/6J mice have about 100 million cells in their brains). My thanks to Richelle Strom, Guomin Zhou, and Dan Goldowitz for allowing me use some of our unpublished data in this review. Richelle C. Strom's Ph.D. dissertation (July 1999) on the quantitative genetic analysis of brain weight and retinal ganglion cell number in mice. Seven chapters containing lots of great new data on QTLs that modulate brain weight and neuron number.
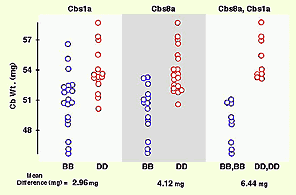
Reprint of a paper by Lu Lu and colleagues published in The Journal of Neuroscience (May 2001). We discovered QTLs on chromosomes 1 and 5 that modulate size and neuron number in the mouse hippocampus. In addition to the mapping, this paper contains extensive morphometric data and cell counts on hippocampus and its components as a function of age, sex, and brain weight. We found an interesting gain in hippocampal weight with age that may be related to continuous adult neurogenesis. We see a similar increase in the olfactory bulbs, but not in the cerebellum. The olfactory bulbs are a great neural system for quantitative genetic analysis (they are modular, discrete, and easy to dissect). Here we describe a set of four QTLs that control the weight of the olfactory bulbs. This paper also details differences in bulb as a function of age, sex, and brain weight. Published in Behavior Genetics, 2001. Reprint of a paper in The Journal of Neuroscience by David C. Airey and colleagues. This study covers both the total size of the cerebellum and the fractional volume of different cerebellar compartments. David and I mapped five QTLs with relatively intense effects on cerebellar size in a cross between C57BL/6J and DBA/2J and in BXD recombinant inbred strains. The figure above shows the effects of two of these QTLs—singly and jointly—on cerebellar weight in the BXD RI strains. 
A stereological and genetic analysis of the caudate nucleus by Rosen and Williams published in BMC Neuroscience (2001; available online at www.biomedcentral.com/browse/biology). While there is considerable strain variation in caudate volume, there is less variation in the number of caudate neurons (mostly medium spiny neurons). Using an F2 intercross we were able to map two QTLs that independently control striatal volume (Chr 10) and striatal neuron number (Chr 19). This paper describes a new technique to exploit consomic mice (also known as chromosome substitution strains) to increase the sensitivity of a recessive mutagenesis screen by restricting analysis to entire litters of nearly isogenic mice. The method is sensitive enough to detect QTLs and weak alleles. The consomic mutatgenesis screen is being used by the Tenneessee Mouse Genomics Consortium to generate and map CNS and behavioral mutations on chromosome 19. This paper was published in Mammalian Genome (1999). A paper on the genetic basis of variation in the eye and retina published in Seminars in Cell & Developmental Biology (1998) 9:249-255. We explain how QTL analysis can be used to detect and characterize genes that modulate the architecture of the eye and retina. We include original data on the genetic control of (1) eye weight, (2) numbers of horizontal cells, and (3) numbers of retinal ganglion cells. This paper was coauthored with Richelle Strom, Guomin Zhou, and Yan Zhen.
An updated and extended version of a paper published in The Journal of Neuroscience (1996, pdf). This paper address the role of genes and envrionmental factors in controlling the size of cell populations in the central nervous system. Quantitative electron microscopy was used to count neurons in retinas of over 450 animals. We measured effects of sex and age, heritability of differences in neuron populations, and the number of genes responsible for the substantial differences among strains of mice. Updated May 30, 1998. This paper follows up on the previous article and was published in The Journal of Neuroscience (1998, pdf). Our aim in this work was to map genes that produce large differences in numbers of neurons in the retina. We succeeded in mapping a gene locus called Neuron Number Control 1 or Nnc1, on chromosome 11 between Hoxb and Krt1. There are several great candidate genes in this region, particularly the thyroid hormone alpha receptor. Nnc1 is the first gene locus known to control normal variation in neuron number in a vertebrate. Our third paper in this series on the genetic control of the retinal ganglion cell population in mice. By estimating total cell production in neonates from 10 different inbred strains of mice, Richelle Strom was able to determine that the distinct bimodality of strain averages is caused by differences in cell production. This paper demonstrates that Nnc1 modulates cell production and must be expressed before birth. However, there are some very interesting differences in the severity of cell death among strains that would be worth following up on.
|
|
Neurogenetics at University of Tennessee Health Science Center
| Top of Page |
Home Page | Genome DBs | Phenome DBs | Publications | People & Associates
Mouse Brain Library | Related Sites | Complextrait.org